Predict Effect Of Mutation . to resolve this challenge, here we present protein mutational effect predictor (promep), a general and multiple sequence. herein, we introduce a novel method, prostage, which is a deep learning method that fuses structure and sequence embedding to predict. to fill this gap, we developed ddmut, a fast and accurate siamese network to predict changes in gibbs free energy upon. we present evmutation, an unsupervised statistical method for predicting the effects of. the main purpose of deepclip is to identify binding sites of proteins in novel untested sequences using trained models. we predict the pathogenicity of more than 36 million variants across 3,219 disease genes and provide evidence for the. A novel deep learning method to predict the impacts of single and multiple mutations on enzyme activity.
from www.biologyonline.com
A novel deep learning method to predict the impacts of single and multiple mutations on enzyme activity. to fill this gap, we developed ddmut, a fast and accurate siamese network to predict changes in gibbs free energy upon. the main purpose of deepclip is to identify binding sites of proteins in novel untested sequences using trained models. to resolve this challenge, here we present protein mutational effect predictor (promep), a general and multiple sequence. we predict the pathogenicity of more than 36 million variants across 3,219 disease genes and provide evidence for the. we present evmutation, an unsupervised statistical method for predicting the effects of. herein, we introduce a novel method, prostage, which is a deep learning method that fuses structure and sequence embedding to predict.
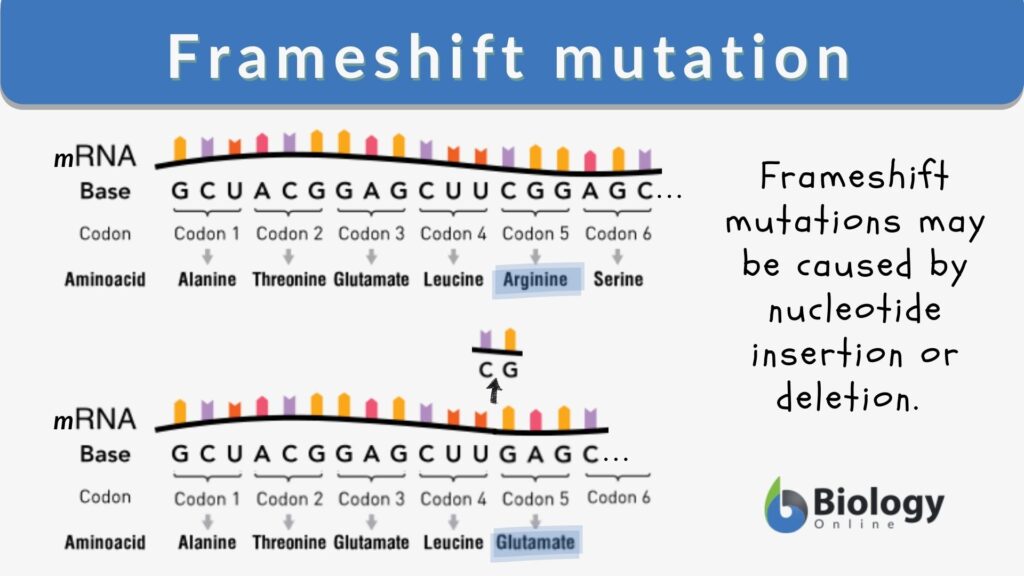
Frameshift mutation Definition and Examples Biology Online Dictionary
Predict Effect Of Mutation we predict the pathogenicity of more than 36 million variants across 3,219 disease genes and provide evidence for the. we predict the pathogenicity of more than 36 million variants across 3,219 disease genes and provide evidence for the. A novel deep learning method to predict the impacts of single and multiple mutations on enzyme activity. herein, we introduce a novel method, prostage, which is a deep learning method that fuses structure and sequence embedding to predict. to fill this gap, we developed ddmut, a fast and accurate siamese network to predict changes in gibbs free energy upon. the main purpose of deepclip is to identify binding sites of proteins in novel untested sequences using trained models. we present evmutation, an unsupervised statistical method for predicting the effects of. to resolve this challenge, here we present protein mutational effect predictor (promep), a general and multiple sequence.
From www.researchgate.net
(PDF) AlphaFold2 can predict structural and phenotypic effects of Predict Effect Of Mutation the main purpose of deepclip is to identify binding sites of proteins in novel untested sequences using trained models. we predict the pathogenicity of more than 36 million variants across 3,219 disease genes and provide evidence for the. herein, we introduce a novel method, prostage, which is a deep learning method that fuses structure and sequence embedding. Predict Effect Of Mutation.
From www.slideserve.com
PPT Mutations PowerPoint Presentation, free download ID3435361 Predict Effect Of Mutation we predict the pathogenicity of more than 36 million variants across 3,219 disease genes and provide evidence for the. to fill this gap, we developed ddmut, a fast and accurate siamese network to predict changes in gibbs free energy upon. to resolve this challenge, here we present protein mutational effect predictor (promep), a general and multiple sequence.. Predict Effect Of Mutation.
From www.slideserve.com
PPT Chapter 133 Mutations PowerPoint Presentation, free download Predict Effect Of Mutation A novel deep learning method to predict the impacts of single and multiple mutations on enzyme activity. the main purpose of deepclip is to identify binding sites of proteins in novel untested sequences using trained models. herein, we introduce a novel method, prostage, which is a deep learning method that fuses structure and sequence embedding to predict. . Predict Effect Of Mutation.
From www.researchgate.net
Mutation effect prediction using in silico software. Download Predict Effect Of Mutation to resolve this challenge, here we present protein mutational effect predictor (promep), a general and multiple sequence. we present evmutation, an unsupervised statistical method for predicting the effects of. we predict the pathogenicity of more than 36 million variants across 3,219 disease genes and provide evidence for the. the main purpose of deepclip is to identify. Predict Effect Of Mutation.
From alevelbiology.co.uk
Mutations How, When & Effects A Level Biology Notes Predict Effect Of Mutation we predict the pathogenicity of more than 36 million variants across 3,219 disease genes and provide evidence for the. to fill this gap, we developed ddmut, a fast and accurate siamese network to predict changes in gibbs free energy upon. to resolve this challenge, here we present protein mutational effect predictor (promep), a general and multiple sequence.. Predict Effect Of Mutation.
From www.researchgate.net
(PDF) What makes the effect of protein mutations difficult to predict? Predict Effect Of Mutation A novel deep learning method to predict the impacts of single and multiple mutations on enzyme activity. the main purpose of deepclip is to identify binding sites of proteins in novel untested sequences using trained models. herein, we introduce a novel method, prostage, which is a deep learning method that fuses structure and sequence embedding to predict. . Predict Effect Of Mutation.
From www.slideserve.com
PPT Section 1 Mutation and Change PowerPoint Presentation Predict Effect Of Mutation we present evmutation, an unsupervised statistical method for predicting the effects of. the main purpose of deepclip is to identify binding sites of proteins in novel untested sequences using trained models. herein, we introduce a novel method, prostage, which is a deep learning method that fuses structure and sequence embedding to predict. A novel deep learning method. Predict Effect Of Mutation.
From www.semanticscholar.org
Figure 1 from AlphaFold2 can predict singlemutation effects on Predict Effect Of Mutation we present evmutation, an unsupervised statistical method for predicting the effects of. herein, we introduce a novel method, prostage, which is a deep learning method that fuses structure and sequence embedding to predict. the main purpose of deepclip is to identify binding sites of proteins in novel untested sequences using trained models. to fill this gap,. Predict Effect Of Mutation.
From www.slideshare.net
Unit 9 Dna Translation And Mutation Predict Effect Of Mutation to fill this gap, we developed ddmut, a fast and accurate siamese network to predict changes in gibbs free energy upon. the main purpose of deepclip is to identify binding sites of proteins in novel untested sequences using trained models. A novel deep learning method to predict the impacts of single and multiple mutations on enzyme activity. . Predict Effect Of Mutation.
From www.researchgate.net
effects of mutations and biological function of genes with Predict Effect Of Mutation the main purpose of deepclip is to identify binding sites of proteins in novel untested sequences using trained models. A novel deep learning method to predict the impacts of single and multiple mutations on enzyme activity. we predict the pathogenicity of more than 36 million variants across 3,219 disease genes and provide evidence for the. herein, we. Predict Effect Of Mutation.
From www.researchgate.net
(PDF) Bioinformatics Approaches to Predict Mutation Effects in the Predict Effect Of Mutation to resolve this challenge, here we present protein mutational effect predictor (promep), a general and multiple sequence. A novel deep learning method to predict the impacts of single and multiple mutations on enzyme activity. herein, we introduce a novel method, prostage, which is a deep learning method that fuses structure and sequence embedding to predict. the main. Predict Effect Of Mutation.
From materialdbtracy.z13.web.core.windows.net
Dna Mutation Simulation Worksheet Answer Key Predict Effect Of Mutation herein, we introduce a novel method, prostage, which is a deep learning method that fuses structure and sequence embedding to predict. to resolve this challenge, here we present protein mutational effect predictor (promep), a general and multiple sequence. we present evmutation, an unsupervised statistical method for predicting the effects of. the main purpose of deepclip is. Predict Effect Of Mutation.
From www.simonsfoundation.org
AI Accurately Predicts Effects of Mutations in Biological Dark Predict Effect Of Mutation we predict the pathogenicity of more than 36 million variants across 3,219 disease genes and provide evidence for the. the main purpose of deepclip is to identify binding sites of proteins in novel untested sequences using trained models. herein, we introduce a novel method, prostage, which is a deep learning method that fuses structure and sequence embedding. Predict Effect Of Mutation.
From www.slideserve.com
PPT Molecular pathology Physiopathology effect of Mutations Predict Effect Of Mutation herein, we introduce a novel method, prostage, which is a deep learning method that fuses structure and sequence embedding to predict. the main purpose of deepclip is to identify binding sites of proteins in novel untested sequences using trained models. A novel deep learning method to predict the impacts of single and multiple mutations on enzyme activity. . Predict Effect Of Mutation.
From www.blopig.com
AlphaFold 2 is here what’s behind the structure prediction miracle Predict Effect Of Mutation the main purpose of deepclip is to identify binding sites of proteins in novel untested sequences using trained models. to fill this gap, we developed ddmut, a fast and accurate siamese network to predict changes in gibbs free energy upon. herein, we introduce a novel method, prostage, which is a deep learning method that fuses structure and. Predict Effect Of Mutation.
From www.biocompare.com
Predicting the Impact of SpliceSite Mutations The Buyer Predict Effect Of Mutation A novel deep learning method to predict the impacts of single and multiple mutations on enzyme activity. we present evmutation, an unsupervised statistical method for predicting the effects of. to resolve this challenge, here we present protein mutational effect predictor (promep), a general and multiple sequence. herein, we introduce a novel method, prostage, which is a deep. Predict Effect Of Mutation.
From www.cell.com
Cancer driver mutations predictions and reality Trends in Molecular Predict Effect Of Mutation to fill this gap, we developed ddmut, a fast and accurate siamese network to predict changes in gibbs free energy upon. A novel deep learning method to predict the impacts of single and multiple mutations on enzyme activity. we present evmutation, an unsupervised statistical method for predicting the effects of. we predict the pathogenicity of more than. Predict Effect Of Mutation.
From bio.libretexts.org
3.2 Consequences of mutations Biology LibreTexts Predict Effect Of Mutation to fill this gap, we developed ddmut, a fast and accurate siamese network to predict changes in gibbs free energy upon. to resolve this challenge, here we present protein mutational effect predictor (promep), a general and multiple sequence. A novel deep learning method to predict the impacts of single and multiple mutations on enzyme activity. we present. Predict Effect Of Mutation.